The Laboratory of Metabolomics at the Institute of Physiology of the Czech Academy of Sciences (IPHYS CAS) provides fee-based services for liquid chromatography–mass spectrometry-based (LC−MS) analysis of polar metabolites, complex lipids, and exposome compounds (including drugs) in biological materials such as plasma, serum, tissues, and cells. These compounds are usually in the range of 50 to 2000 Da and their concentrations may span several orders of magnitude. The laboratory also offers analysis of investigational medicinal products (IMPs) and pharmaceuticals.
The laboratory is equipped with three state-of-the-art LC-MS systems (Thermo Vanquish UHPLC/Q Exactive Plus, Thermo Vanquish UHPLC/Orbitrap Exploris 480*, and Dionex UltiMate 3000 UHPLC/SCIEX QTRAP 5500), a GC-MS system (Agilent 990 GC/Leco Pegasus BT2*), and an Agilent Bravo Automated Liquid Handling Platform*.
* Supported by the National Institute for Research of Metabolic and Cardiovascular Diseases (Programme EXCELES, ID Project No. LX22NPO5104)—Funded by the European Union–Next Generation EU.
 MetaboAtlas21
MetaboAtlas21
 GTTAtlas
GTTAtlas
LIMeX
UNTARGETED METABOLOMICS, LIPIDOMICS, AND EXPOSOMICS
LIMeX (LIpids, Metabolites, and eXposome compounds) LC−MS workflow is used for the untargeted analysis of complex lipids, polar metabolites, and exposome compounds (such as drugs).

Bi-phase extraction method using methanol, methyl-terc butyl ether (MTBE), and water provides two phases: one containing polar metabolites (aka “metabolomics”) and different exposome compounds such as drugs (aka “exposomics”), and the other with complex lipids (aka “lipidomics”). During untargeted analyses, all detectable compounds are acquired using validated LC–MS methods.
Since no single LC–MS method can cover the full metabolome/lipidome/exposome, we use different chromatography modes (reversed-phase liquid chromatography, RPLC and hydrophilic interaction chromatography, HILIC) and electrospray ionization modes to comprehensively characterize these isolated fractions.
A high-resolution mass spectrometer Thermo Q Exactive Plus coupled with a liquid chromatograph (Thermo Vanquish) is routinely used for LIMeX workflow. The mass spectrometer collects full scan MS1 data as well as data-dependent MS/MS spectra for all samples. While MS1 data is used for quantification, MS/MS spectra are used for compound annotation using MS/MS library search or for compound confirmation. The acquired data sets are processed using MS-DIAL 4 software, including annotation of complex lipids, polar metabolites, and other detected compounds using in-house, open-source as well as commercial MS/MS spectral libraries (NIST20, MassBank, MoNA).
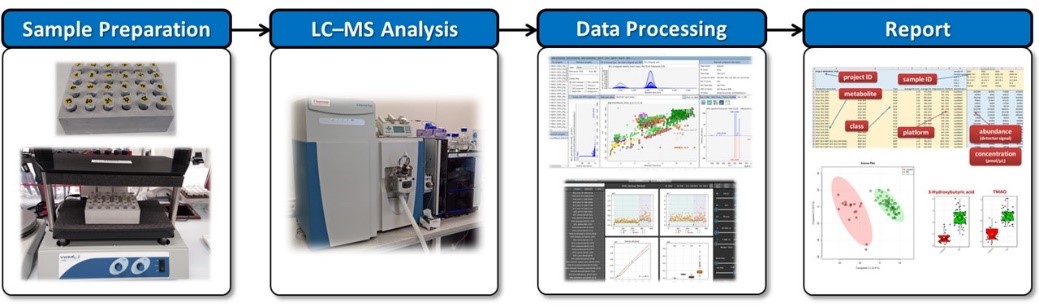
LIMeX workflow for untargeted and targeted analysis of complex lipids, polar metabolites, and exposome compounds.
For human cohort studies, we usually report over 500 complex lipids, 100 polar metabolites, and tens of exposome compounds (mainly food components and drugs) using LIMeX−4D (i.e., 4 LC−MS platforms).
All compounds are quantified as peak heights and reported in “arbitrary units”. These values are used as inputs for one-dimensional or multidimensional statistical methods (e.g., t-test, fold-change, heatmap, principal component analysis, partial least squares-discriminant analysis).
The LIMeX workflow uses over 60 different internal standards, which can be used for estimations of absolute concentrations for various metabolites detected in plasma/serum and tissues.
Lipid mediators
TARGETED ANALYSIS OF LIPID MEDIATORS
Targeted quantitative LC−MS/MS analysis is available for specific low-abundant lipid mediators (eicosanoids, endocannabinoids, and fatty acid esters of hydroxy fatty acids (FAHFA)) and steroids.
The sample preparation for these low-abundant compounds is more demanding and solid-phase extraction (SPE) is involved in the workflow to remove high-abundant compounds (e.g., phospholipids).
A quadrupole/linear ion trap mass spectrometer (SCIEX QTRAP 5500) operated in multiple reaction monitoring (MRM) coupled with a liquid chromatograph (Dionex/Thermo Ultimate 3000 RSLC) is used for analysis.
Fluxomics
TARGETED FLUXOMICS
Targeted LC−MS methods are used to measure metabolites labeled with stable isotopes using 13C/2H glucose and 2H2O tracers. LIMeX protocol is used for sample extraction followed by LC−MS analysis in which case the mass spectrometer (Q Exactive Plus) is operated in ultra-high-resolution mode (140,000 FWHM at m/z 200).
Theoretical isotopologues are calculated for annotated metabolites in Python and instrumental files are processed through MRMPROBS software. Peak heights are adjusted for the natural abundance of elements and tracer purity using IsoCor.
Pharma

TARGETED ANALYSIS OF IMP & PHARMACEUTICALS
Targeted LC−MS methods are used for Good Laboratory Practice (GLP) and non-GLP analysis of investigational medicinal products (IMP) and pharmaceutical compounds, including also potential drug candidates within absorption, distribution, metabolism and excretion (ADME) studies.
The lab is GLP certified for analytical and clinical chemistry testing.
For GLP studies, the following three steps are essential:
A high-resolution mass spectrometer Thermo Q Exactive Plus coupled with a liquid chromatograph (Thermo Vanquish) is routinely used for the analysis of pharmaceutical compounds.
Submit samples

To submit your samples, please use the following submission form. All platforms have a minimum sample requirement of 10 samples. The form also contains information on a minimum amount or volume requirement per sample for each platform.
Send this form to Dr. Tomáš Čajka and deliver your samples to:
- Institute of Physiology CAS
- Laboratory of Metabolomics, room E-216
- Videnska 1083, Prague, 14220
- Czech Republic
After receiving your samples, a unique project ID is generated and is used within all platforms, for the final report, and data archiving.
People

doc. Ing. Tomáš Čajka, Ph.D. Head of the Laboratory
Ing. Jiří Hricko Research Assistant LC–MS, GLP Mgr. Michaela Paučová Research Assistant LC–MS, GLP Mgr. Tatyana Kobets, Ph.D. Research Assistant statistics, bioinformatics Ing. Michaela Nováková Research Assistant statistics, bioinformatics Bc. Aleksandra Shumilova Research Assistant statistics, bioinformatics Mgr. Petra Zedníková (maternity leave) Research Assistant
Publications

The laboratory has provided metabolomics and lipidomics data for several projects worldwide. Below is the list of papers published in peer-reviewed journals.
- Brejchova et al., Metabolism 166 (2025) 156157 (doi: 10.1016/j.metabol.2025.156157)
- Arora et al., Acta Physiol 241 (2025) e14278 (doi: 10.1111/apha.14278)
- Melenovský et al., Clinical Diabetes 43 (2025) 23-32cd240003 (doi: 10.2337/cd24-0003)
- Pecina et al., Commun Biol 7 (2024) 1116 (doi: 10.1038/s42003-024-06819-w)
- Riecan et al., Biochim. Biophys. Acta, Mol. Cell Biol. Lipids 1869 (2024) 159543 (doi: 10.1016/j.bbalip.2024.159543)
- Šiklová et al., Obesity 32 (2024) 547 (doi: 10.1002/oby.23969)
- Rakusanova & Cajka, Trends Anal. Chem. 180 (2024) 117940. (doi: 10.1016/j.trac.2024.117940)
- Rakusanova & Cajka, Physiol. Res. 73 (2024) S165 (doi: 10.33549/physiolres.935443)
- Cajka, Curr Opin Food Sci 58 (2024) 101201 (doi: 10.1016/j.cofs.2024.101201)
- Kasperova et al., Cardiovasc Diabetol 23 (2024) 223 (doi: 10.1186/s12933-024-02298-9)
- Balcaen et al., J Lipid Res 65 (2024) 100572 (doi: 10.1016/j.jlr.2024.100572)
- Hořejší et al., J Biol Chem 300 (2024) 107154 (doi: 10.1016/j.jbc.2024.107154)
- Cajka et al., Int J Mol Sci 25 (2024) 2899 (doi: 10.3390/ijms25052899)
- Castillo et al., BioFactors 49 (2023) 1022 (doi: 10.1002/biof.1974)
- Benova et al., Commun Biol 6 (2023) 1043 (doi: 10.1038/s42003-023-05407-8)
- Ferencakova et al., Front Cell Dev Biol 11 (2023) 1255823 (doi: 10.3389/fcell.2023.1255823)
- Cajka et al., Metabolites 13 (2023) 966 (doi: 10.3390/metabo13090966)
- Horakova et al., Front Endocrinol 14 (2023) 1205703 (doi: 10.3389/fendo.2023.1205703)
- Hricko et al., Antioxidants 12 (2023) 986 (doi: 10.3390/antiox12050986)
- Harant et al., Biophys Chem 296 (2023) 106989 (doi: 10.1016/j.bpc.2023.106989)
- Janovska et al., Mol Metab 369 (2023) 101683 (doi: 10.1016/j.molmet.2023.101683)
- Cajka et al., Int J Mol Sci 324 (2023) 1987 (doi: 10.3390/ijms24031987)
- Castillo et al., Mol Nutr Food Res 366 (2022) 2200204 (doi: 10.1002/mnfr.202200204)
- Martínez-Ramírez et al., LWT - Food Sci Technol 3173 (2022) 114311 (doi: 10.1016/j.lwt.2022.114311)
- Paluchova et al., Free Radic Biol Med 3193 (2022) 787 (doi: 10.1016/j.freeradbiomed.2022.11.015)
- Benova et al., Mol Metab 365 (2022) 101598 (doi: 10.1016/j.molmet.2022.101598)
- Brejchova et al., Food Chem 388 (2022) 132983 (doi: 10.1016/j.foodchem.2022.132983)
- Pruchova et al., Antioxidants 11 (2022) 198 (doi: 10.3390/antiox11020198)
- Stemberkova-Hubackova et al., Clin Transl Med 12 (2022) e645 (doi: 10.1002/ctm2.645)
- Jacome-Sosa et al., Commun Biol 4 (2021) 1247 (doi: 10.1038/s42003-021-02765-z)
- Lopes et al., Cell Rep 37 (2021) 109833 (doi: 10.1016/j.celrep.2021.109833)
- Spackova et al., Cancers 13 (2021) 1709 (doi: 10.3390/cancers13071709)
- Danhelovska et al., Int J Mol Sci 22 (2021) 7270 (doi: 10.3390/ijms22147270)
- Koc et al., Sci Rep 11 (2021) 8171 (doi: 10.1038/s41598-021-87494-3)
- Sistilli et al., Nutrients 13 (2021) 437 (doi: 10.3390/nu13020437)
- Brejchova et al., Proc Natl Acad Sci USA 118 (2021) e2020999118 (doi: 10.1073/pnas.2020999118)
- Janovska et al., J Cachexia Sarcopenia Muscle 11 (2020) 1614 (doi: 10.1002/jcsm.12631)
- Bardova et al., Nutrients 12 (2020) 3737 (doi: 10.3390/nu12123737)
- Tsugawa et al., Nat Biotechnol 38 (2020) 1159–1163 (doi: 10.1038/s41587-020-0531-2)
- Smolkova et al., Sci Rep 10 (2020) 8677 (doi: 10.1038/s41598-020-65351-z)
- Paluchova et al., Mol Nutr Food Res 64 (2020) 1901238 (doi: 10.1002/mnfr.201901238)
- Benlebna et al., J Nutr Biochem 79 (2020) 108361 (doi: 10.1016/j.jnutbio.2020.108361)
- Paluchova et al., Diabetes 69 (2020) 300–312 (doi: 10.2337/db19-0494)
- Brezinova et al., Biochim Biophys Acta Mol Cell Biol Lipids 1865 (2020) 158576 (doi: 10.1016/j.bbalip.2019.158576)
- Angelisová et al., Biochim Biophys Acta Biomembr 1861 (2019) 130–141 (doi: 10.1016/j.bbamem.2018.08.006)
- Sládek et al., Biochim Biophys Acta Mol Cell Biol Lipids 1864 (2019) 158533 (doi: 10.1016/j.bbalip.2019.158533)

 CZECH
CZECH